PRESENTATION

Mélody MERLE
Doctorante - PhD Student
Tour 13/23 - 5ème étage, bureau 527 (campus Jussieu)
merle(at)lptmc.jussieu.fr
melody.merle(at)gmail.com
Mots clés : pattern formation, Ising asymétrique, réseau de régulation génétique, développement embryonnaire, développement de la Drosophile
Key words : pattern formation, asymmetric Ising model, genetic regulation network, embryonic development, Drosophila development
Bref CV:
- 2016-aujourd'hui : Thèse en biophysique théorique au LPTMC - Modeling the early stage development of Drosophila by an Ising-based model of genetic regulation network.
including: aug2017-dec2017: Visite au Thomas Gregor Lab à Princeton, NJ, USA - Imaging of proteins concentration by immunofluorescence in the Drosophila embryo
- 2015-2016 : Master 2 Systèmes Biologiques et Concepts Physique (SBCP) - Cours Génétique et Epigénétique à l'Institut Pasteur
- 2012-2016 : ESPCI 131ème promotion - spécialité Biotechnologies
Link to complete CV
RECHERCHE
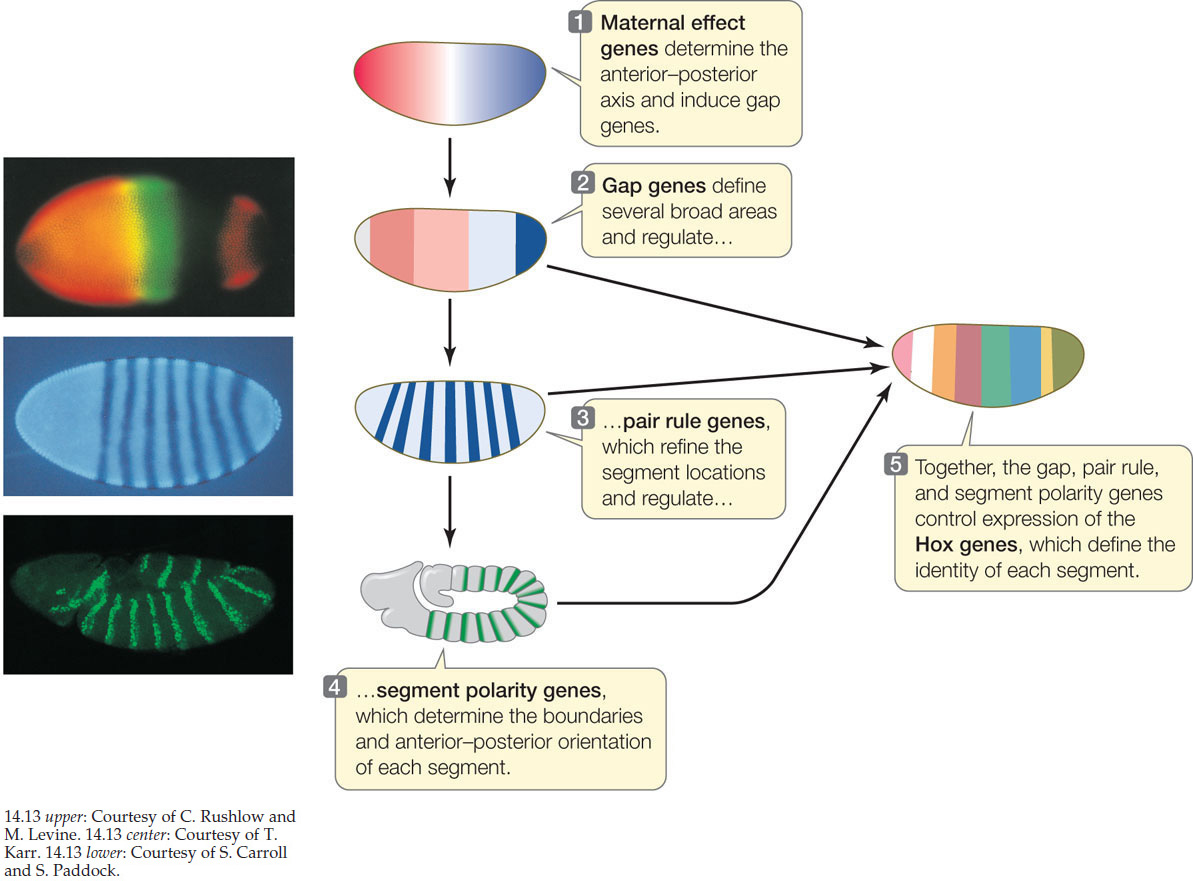
Development biology is the study of how multicellular organims grow and form, starting from an unique cell. Pattern formation is an essential concept in developmental biology as it sets up the body plan and guides initially equivalent cells toward their differentiation in various form and functions. The mechanisms of pattern formation are ensured mainly by genes and their complex regulation mechansims.
In the Drosophila embryo, the antero-posterior axis is patterned by segmentation. This phenomena starts early during embryonic development, before gastrulation. Each segment formed at this stage correspond to a segment of the adult fly (through the Hox genes family). The first genes involved in the segmentation regulate each other hierarchically : maternal effect genes, gap genes, pair-rule gene and segment polarity genes.
At this stage of development, the embryo is consituted of around 6000 nuclei distributed along the embryo surface. In particular, the embryo is a unique cell with multiple nuclei (syncytium). It allows rapid interactions between nucleis as there is no cellular membrane.
Inspired by this example, we derived from the Ising model, a classical statistical physics model for magnetism, a model of genetic regulation network in multicellular organims. To each site of a lattice are associated several "spin", representing genes, taking discrete values 0/1. These genes locally interact with each other through an interaction constant matrix J (where Jab represents the effect of gene A on gene B). Maternal effect genes are assimilated to an external magnetic field. Simulations are made based on the method for asymmetric Ising models, using a Monte Carlo Markov Chain algorithm.
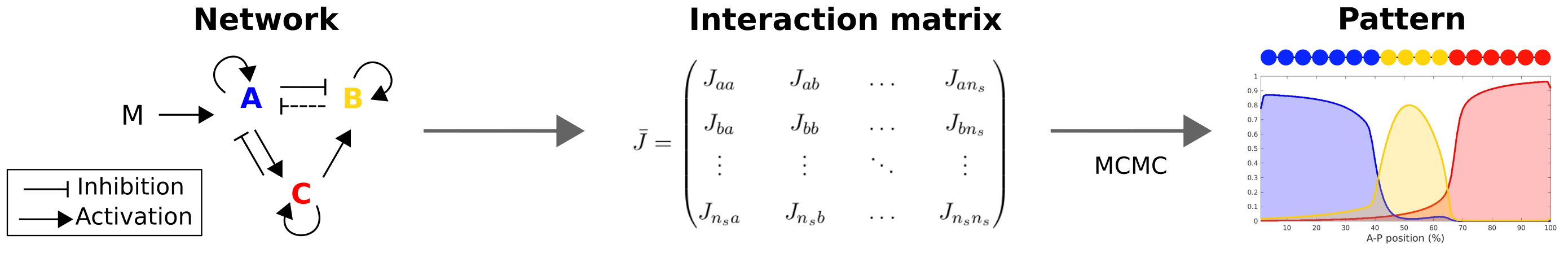
The aim is to discover novel aspects of pattern formation using this new model and to quantify and compare different patterning strategies.
PUBLICATIONS
Under review:
Turing-like pattern in asymmetric dynamic Ising model
M.Merle, L.Messio, J.Mozziconacci
https://arxiv.org/abs/1903.08506 (march 2019)
The 3D genome encodes gene regulation during development
J.Mozziconacci, M.Merle, A.Lesne (may 2019)
ENSEIGNEMENT
TP et Colles : L1 UE Energie et Entrope (1P003) - Jussieu (2017-2019)
Colles : Préparation aux oraux en PC*, lycée Janson de Sailly (2015 et 2016)




